Difference between revisions of "Thematic Examples"
| Line 31: | Line 31: | ||
$ xcrysden --pwi si.scf.david.in | $ xcrysden --pwi si.scf.david.in | ||
</code> | </code> | ||
| − | Which gives us this structure visualization: | + | Which gives us this structure visualization: |
| + | |||
[[File:Si-2x2x2.png]] | [[File:Si-2x2x2.png]] | ||
| + | |||
Showing the typical diamond like tetrahedral structure. | Showing the typical diamond like tetrahedral structure. | ||
We now call '''Quantum Espresso''' and run the three steps, this can be performed in one job for simple short calculations, for longer calculations the three steps can be separated into three jobs. Let us create the three submission scripts. | We now call '''Quantum Espresso''' and run the three steps, this can be performed in one job for simple short calculations, for longer calculations the three steps can be separated into three jobs. Let us create the three submission scripts. | ||
Revision as of 10:30, 9 May 2025
Contents
Thematic Examples From Computer Science, Engineering and Computational Modeling and Material Science
We will work through examples based on the following codes:
- Quantum Espresso (CMMS)
- YAMBO (CMMS)
- GROMACS (CMMS)
- Tensorflow (CS)
- PyTorch (CS)
- FluidX3D (Engineering)
CMMS Thematic Area
Gromacs Tutorial
Quantum Espresso Tutorial
Computing The Bandstructure of Silicon
In this tutorial, we will calculate the band structure of bulk Silicon.
Silicon is a semi-conductor with a small gap. The calculation will proceed through three broad steps, starting from the
self consistent calculation, the non-self consistent calculation, the bands calculation, and finally the post processing.
The files required for this exercise can be retreived as follows:
$ cd ~/localscratch/ $ mkdir examples $ cd examples/ $ git clone https://github.com/Materials-Modelling-Group/training-examples.git $ cd training-examples/ $ ls si.band.david.in si.nscf.david.in si.plotbands.in si.scf.david.in
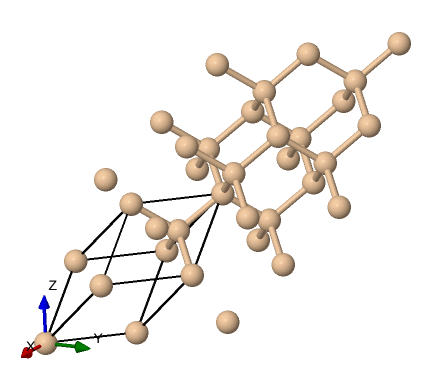
We can visualize the structure using tools like VESTA or Xcrysden, using the input file that specifies the structure si.scf.david.in . This can be done on your computer once you install Xcrysden and copy the input to your device as such:
$ xcrysden --pwi si.scf.david.in
Which gives us this structure visualization:
Showing the typical diamond like tetrahedral structure.
We now call Quantum Espresso and run the three steps, this can be performed in one job for simple short calculations, for longer calculations the three steps can be separated into three jobs. Let us create the three submission scripts.
#!/bin/bash #SBATCH --job-name=si_scf_normal #SBATCH --partition=gpu1 #SBATCH --gpus=1 #SBATCH --ntasks-per-node=1 #SBATCH --time=00:10:00 #SBATCH --output=si_scf_%j.out #SBATCH --error=si_scf_%j.err ##Load necessary modules module load applications/gpu/qespresso/7.3.1 ##Set environment variables for QE cd $HOME/localscratch/examples/training-examples ##Run QE with GPU support mpirun -np 1 pw.x -in si.scf.david.in > scf.out
put this in a file named 1scf.job and we submit the same:
$ sbatch 1scf.job
Once this has run, we will have some generated outputs, including wavefunctions and the text file scf.out.
Next we prepare the submission script for the nscf step,
#!/bin/bash #SBATCH --job-name=si_nscf_normal #SBATCH --partition=gpu1 #SBATCH --gpus=1 #SBATCH --ntasks-per-node=1 #SBATCH --time=00:10:00 #SBATCH --output=si_scf_%j.out #SBATCH --error=si_scf_%j.err ##Load necessary modules module load applications/gpu/qespresso/7.3.1 ##Set environment variables for QE cd $HOME/localscratch/examples/training-examples ##Run QE with GPU support mpirun -np 1 pw.x -in si.nscf.david.in > nscf.out
put this in a file named 2nscf.job and we submit the same:
$ sbatch 2nscf.job
Once this nscf step is done, we now compute the band structure, create the submission script for the bands calculation:
#!/bin/bash #SBATCH --job-name=si_bands_normal #SBATCH --partition=gpu1 #SBATCH --gpus=1 #SBATCH --ntasks-per-node=1 #SBATCH --time=00:10:00 #SBATCH --output=si_scf_%j.out #SBATCH --error=si_scf_%j.err ##Load necessary modules module load applications/gpu/qespresso/7.3.1 ##Set environment variables for QE cd $HOME/localscratch/examples/training-examples ##Run QE with GPU support mpirun -np 1 bands -in si.bands.in > si.bands.out
We have the bulk of the computational work done, we now are left with a post processing steps to obtain results, we now use the
plotbands executable for this step:
#!/bin/bash #SBATCH --job-name=si_ppbands_normal #SBATCH --partition=gpu1 #SBATCH --gpus=1 #SBATCH --ntasks-per-node=1 #SBATCH --time=00:10:00 #SBATCH --output=si_scf_%j.out #SBATCH --error=si_scf_%j.err ##Load necessary modules module load applications/gpu/qespresso/7.3.1 ##Set environment variables for QE cd $HOME/localscratch/examples/training-examples ##Run QE with GPU support mpirun -np 1 plotband.x < si.plotband.in
This will produce some new visualization data and a postscript plot, si_bands.ps which should yield an image like this:
CS Thematic Area
TODO
Conda Usage on the cluster
TODO
PyTorch Tutorial
TODO
TensorFlow Tutorial
Engineering Thematic Area
TODO.
FluidX3D Tutorial
TODO
Up: HPC_Usage